
B.C. tenants evicted for landlord's use after refusing large rent increase to take over neighbouring suite
Ashley Dickey and her mother rented part of the same Coquitlam duplex in three different decades under three different landlords.
A team of scientists from India, Sweden and Japan has developed a new diagnostic tool capable of keeping pace with rapidly mutating bacteria and viruses.
At a time when the world has seen first-hand how novel germs – like SARS-CoV-2 and its variants – can disrupt life on a large scale, researchers say their new tool could speed up the discovery of new viral and bacterial variants, helping combat them quickly and effectively.
"Emergence of novel zoonotic infections among the human population has increased the burden on global health-care systems to curb their spread," the authors wrote in a paper published in the peer-reviewed scientific journal ACS Infectious Diseases on Wednesday.
"To meet the evolutionary agility of pathogens, it is essential to revamp the existing diagnostic methods for early detection and characterization of the pathogens at the molecular level."
The tool, a computer program called AutoPLP, builds on the technology polymerase chain reaction (PCR) tests use to detect viruses and bacteria based on their genetic material. PCR tests use nucleic acid probes – short, single-stranded sequences of DNA or RNA coded to match a specific pathogen – to diagnose infections by searching for the matching DNA or RNA associated with the pathogen they're testing for.
Rolling circle amplification (RCA) tests work similarly to PCR tests, but are known for being more sensitive than PCR tests because they use a special, more precise type of nucleic acid probe called a padlock probe (PLP). This makes them useful for detecting mutated genetic sequences, like those in novel viruses and viral variants.
The caveat is, because padlock probes are so precise, it takes more work to pinpoint specific genetic sequences for them to target. On top of that, as a pathogen mutates, its genetic sequence changes as well, forcing scientists to redesign their probes each time.
So researchers Sowmya Ramaswamy Krishnan, Ruben R. G. Soares, Narayanan Madaboosi, and M. Michael Gromiha set out to develop a software program that could keep up with viral and bacterial mutations better than human scientists can.
AutoPLP designs PLPs automatically to match genetically mutating bacteria and viruses, while systematically considering "all the necessary technical parameters at once to make the entire process easier and more robust."
The program can take the genome sequences of similar pathogens as input and run a series of analyses and database searches, producing a set of customized PLP sequences.
The authors tested the tool by having it design probes to detect the rabies virus and drug-resistant strains of Mycobacterium tuberculosis, the bacterium responsible for tuberculosis. The probes designed by AutoPLP worked more quickly, accurately and easily than previous probes developed to detect these pathogens.
The authors hope the tool will hasten the discovery of new viral and bacterial variants and help "combat them rapidly and effectively via precise molecular diagnostics," according to a media release issued on Wednesday.

Ashley Dickey and her mother rented part of the same Coquitlam duplex in three different decades under three different landlords.
A man who fell into a crevasse while leading a backcountry ski group deep in the Canadian Rockies has died.
A new survey by Dalhousie University's Agri-Food Analytics Lab asked Canadians about their food consumption habits amid rising prices.
MPP Sarah Jama was asked to leave the Legislative Assembly of Ontario by House Speaker Ted Arnott on Thursday for wearing a keffiyeh, a garment which has been banned at Queen’s Park.
Charlie Woods failed to advance in a U.S. Open local qualifying event Thursday, shooting a 9-over 81 at Legacy Golf & Tennis Club.
As Donald Trump was running for president in 2016, his old friend at the National Enquirer was scooping up potentially damaging stories about the candidate and paying out tens of thousands of dollars to keep them from the public eye.
After Prime Minister Justin Trudeau said the federal government would still send Canada Carbon Rebate cheques to Saskatchewan residents, despite Saskatchewan Premier Scott Moe's decision to stop collecting the carbon tax on natural gas or home heating, questions were raised about whether other provinces would follow suit. CTV News reached out across the country and here's what we found out.
A Montreal actress, who has previously detailed incidents she had with disgraced Hollywood producer Harvey Weinstein, says a New York Court of Appeals decision overturning his 2020 rape conviction is 'discouraging' but not surprising.
Caleb Williams is heading to the Windy City, aiming to become the franchise quarterback Chicago has sought for decades.
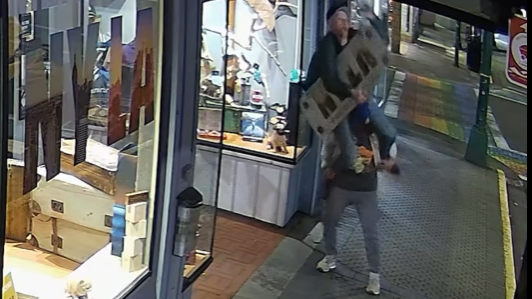
Mounties in Nanaimo, B.C., say two late-night revellers are lucky their allegedly drunken antics weren't reported to police after security cameras captured the men trying to steal a heavy sign from a downtown business.
A property tax bill is perplexing a small townhouse community in Fergus, Ont.
When identical twin sisters Kim and Michelle Krezonoski were invited to compete against some of the world’s most elite female runners at last week’s Boston Marathon, they were in disbelief.
The giant stone statues guarding the Lions Gate Bridge have been dressed in custom Vancouver Canucks jerseys as the NHL playoffs get underway.
A local Oilers fan is hoping to see his team cut through the postseason, so he can cut his hair.
A family from Laval, Que. is looking for answers... and their father's body. He died on vacation in Cuba and authorities sent someone else's body back to Canada.
A former educational assistant is calling attention to the rising violence in Alberta's classrooms.
The federal government says its plan to increase taxes on capital gains is aimed at wealthy Canadians to achieve “tax fairness.”
At 6'8" and 350 pounds, there is nothing typical about UBC offensive lineman Giovanni Manu, who was born in Tonga and went to high school in Pitt Meadows.